Development and application of crystallographic methodologies for the structure determination of molecules of biological or technological interest with different complexity
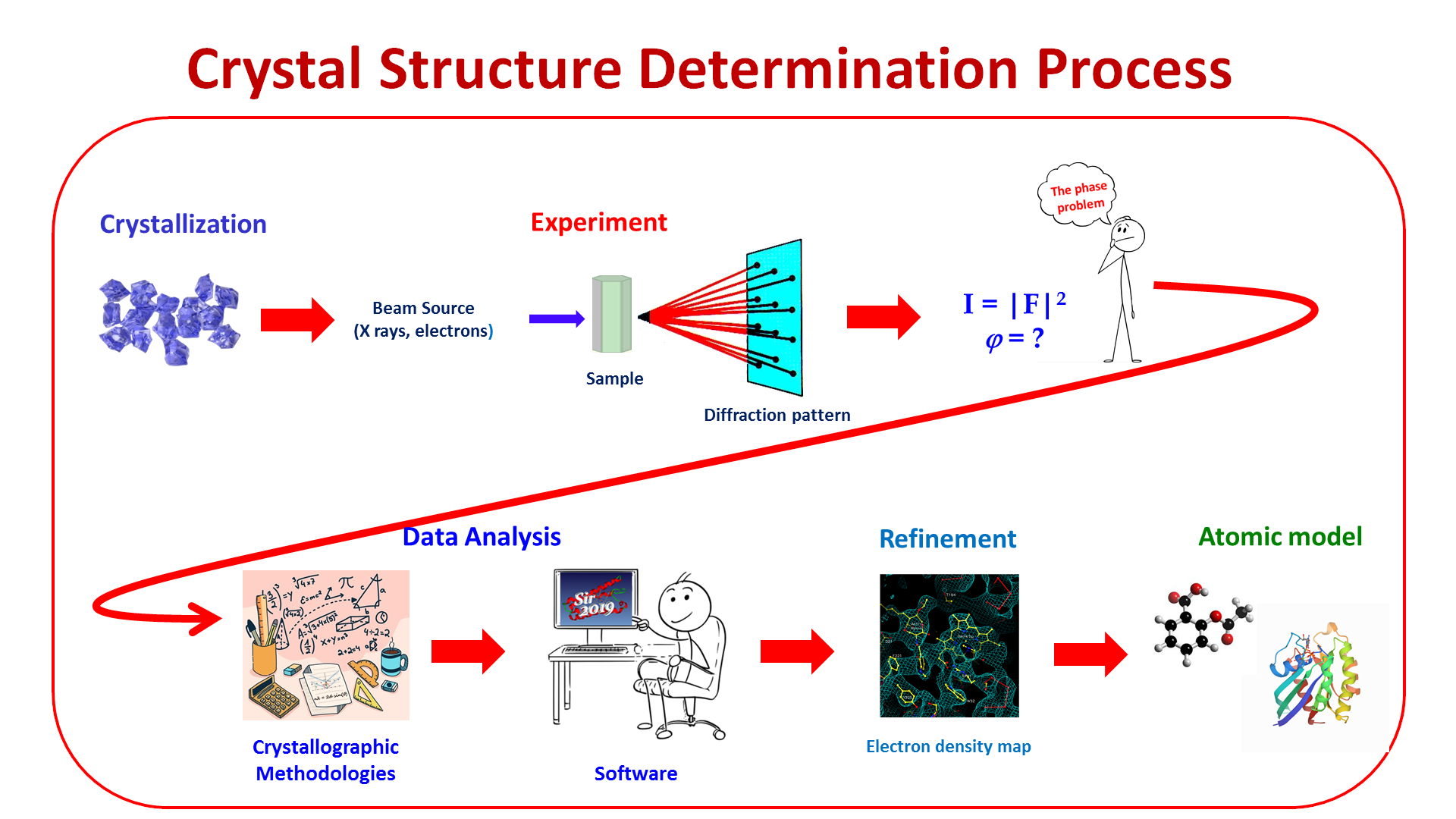
Development and application of computational methods for crystal structure determination of compounds with different nature and complexity, using diffraction data (X-rays or electrons), with special attention to molecules of biological or technological interest.
Crystallographic methodologies are fundamental for studying physical and chemical properties or biological functionalities of compounds with different compositions and complexities (from small molecules to proteins), since they strongly depend on the arrangement of the atoms within the molecules.
The structure solution of small molecules is today considered a routine, even if not trivial, process; on the contrary the structure determination of proteins or nucleic acids, whose data are often incomplete and at low resolution, is still a complicated and not completely automated process.
This research activity is aimed at:
– the development of innovative crystallographic methods for structure solution of crystalline materials with different complexity, especially with regard to implementation of specific pipelines (from phasing to the Automated Model Building) dedicated to macromolecules, based on different approaches: Ab Initio, MAD/SAD and in particular Molecular Replacement (the most used);
– constant updating of the different structure solution algorithms implemented in latest release of the SIR20XX software (produced by the IC researchers), to keep offering to the international community an efficient, flexible, performing and user-friendly program, already widely and profitably used today by thousands of researchers around the world;
– the application of the new procedures to real cases.
– Burla M.C., Carrozzini B., Cascarano G.L., Giacovazzo C. & Polidori G. (2020) – Cyclic automated model building (CAB) applied to nucleic acids – Crystals, 10, 280. Doi: 10.3390/cryst10040280
– Burla M.C., Carrozzini B., Cascarano G.L., Giacovazzo C. & Polidori G. (2020) – How far are we from automatic crystal structure solution via molecular-replacement techniques? – Acta. Cryst., D76, 9-18. Doi: 10.1107/S2059798319015468
– Burla M.C., Carrozzini B., Cascarano G.L., Giacovazzo C. & Polidori G. (2018) – CAB: a cyclic automatic model-building procedure – Acta. Cryst., D74, 1096-1104. Doi: 10.1107/S2059798318013438
– Burla M.C., Caliandro R., Carrozzini B., Cascarano G.L., Cuocci C., Giacovazzo C., Mallamo M., Mazzone A. & Polidori G. (2015) – Crystal structure determination and refinement via SIR2014- Jour. Appl. Cryst., 48, 306-309. Doi: 10.1107/S1600576715001132


