High-resolution structural investigation of protein/ligand interactions
Structural characterization of bio-macromolecules in the presence of pharmacological/biotechnological-interesting compounds to provide useful information for the optimization process of the latter ones as well as the identification of the molecular mechanism underlying their interaction with the macromolecule.
Unveiling the binding between a biological macromolecule and a small molecule is a key step for the optimization process of the small molecule itself, particularly in the case of molecules of pharmaceutical/biotechnological interest. Moreover, in the case the ligand is a natural binder of the macromolecule, describing the interaction between the two molecular entities is useful for the understanding of the biochemical mechanisms in which they are involved.
Obtaining such information is at the heart of our research activity. Our focus is the structural aspect of the interaction that is investigated by high-resolution X-ray-based techniques such as diffraction (MX) and absorption (XAS). By exploiting such techniques, we have investigated macromolecules involved in biochemical processes such as apoptosis, DNA methylation, intra and extra cellular proteolysis and metal homeostasis. The provided high resolution information combined with fragment and structure based molecular design techniques have allowed to develop new inhibitors, synthetic allosteric modulators, molecular antennas for applications in the photovoltaic field as well as for the optimization of proteins of interest in the biotechnology field.
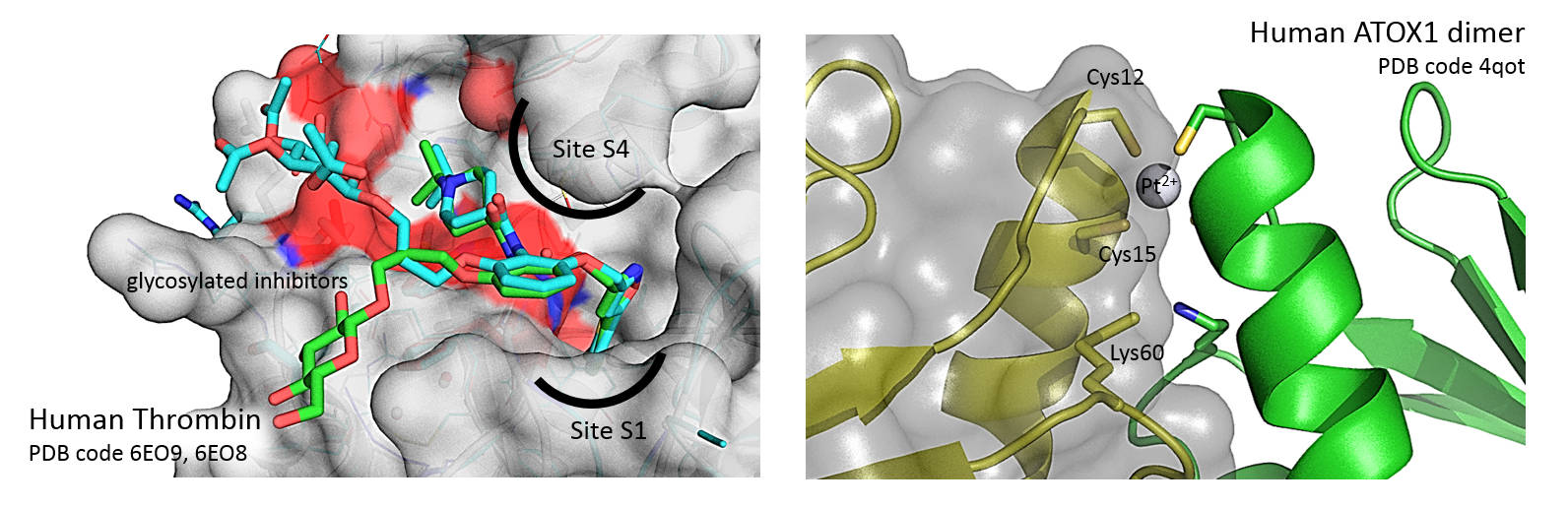
– BD Belviso, A Galliani, A Lasorsa, V Mirabelli, R Caliandro, F Arnesano, and G Natile, Oxaliplatin Binding to Human Copper Chaperone Atox1 and Protein Dimerization, Inorganic Chemistry, 2016, 55 (13), 6563-6573, DOI: 10.1021/acs.inorgchem.6b00750
– BD Belviso, R Caliandro, M de Candia, G Zaetta, G Lopopolo, F Incampo, M Colucci, and CD Altomare, How a β-D-Glucoside Side Chain Enhances Binding Affinity to Thrombin of Inhibitors Bearing 2-Chlorothiophene as P1 Moiety: Crystallography, Fragment Deconstruction Study and Evaluation of Antithrombotic Properties, Journal of Medicinal Chemistry, 2014, 57 (20), 8563-8575, DOI: 10.1021/jm5010754
– BD Belviso, R Caliandro, D Siliqi, V Calderone, F Arnesano, and G Natile, Structure of matrix metalloproteinase-3 with a platinum-based inhibitor, Chemical Communications, 2013,49, 5492-5494, DOI: 10.1039/C3CC41278D
– M Catto, L Pisani, E de la Mora, BD Belviso, GF Mangiatordi, A Pinto, A De Palma, N Denora, R Caliandro, Jacques-Philippe Colletier, Israel Silman, Orazio Nicolotti, and Cosimo Damiano Altomare, Chiral Separation, X-ray Structure and Biological Evaluation of a Potent and Reversible Dual Binding Site AChE Inhibitor, ACS Medicinal Chemistry Letters 2020 11 (5), 869-876, DOI: 10.1021/acsmedchemlett.9b00656


